Abstract
Mini Review
Feature Processing Methods: Recent Advances and Future Trends
Shiying Bai and Lufeng Bai*
Published: 03 April, 2025 | Volume 9 - Issue 1 | Pages: 010-014
This paper reviews recent advances and future trends in feature processing methods within the field of artificial intelligence. With the rapid development of deep learning and big data technologies, feature processing has become essential for enhancing AI model performance. We begin by revisiting traditional feature processing methods, then focus on deep learning-based feature extraction techniques, automated feature engineering, and the application of feature processing in specific domains. The article also analyzes the current research challenges and outlines future development directions, offering structured insights for both researchers and practitioners across disciplines
Read Full Article HTML DOI: 10.29328/journal.jcmei.1001035 Cite this Article Read Full Article PDF
Keywords:
Feature processing; Artificial intelligence; Deep learning; Automated feature engineering; Data preprocessing; Feature selection; Dimensionality reduction
References
- Dhal P, Azad C. A comprehensive survey on feature selection in the various fields of machine learning. Appl Intell. 2022;52(4):4543-4581. Available from: https://link.springer.com/article/10.1007/s10489-021-02550-9
- Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022;28(9):1773-1784. Available from: https://doi.org/10.1038/s41591-022-01981-2
- Alotaibi B, Alotaibi M. A hybrid deep ResNet and inception model for hyperspectral image classification. PFGC J Photogramm Remote Sens Geoinf Sci. 2020;88(6):463-476. Available from: https://link.springer.com/article/10.1007/s41064-020-00124-x
- Peng S, Huang H, Chen W, Zhang L, Fang W. More trainable inception-ResNet for face recognition. Neurocomputing. 2020;411:9-19. Available from: https://doi.org/10.1016/j.neucom.2020.05.022
- Barakbayeva T, Demirci FM. Fully automatic CNN design with inception and ResNet blocks. Neural Comput Appl. 2023;35(2):1569-1580. Available from: http://dx.doi.org/10.1007/s00521-022-07700-9
- Sherstinsky A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network. Physica D Nonlinear Phenom. 2020;404:132306. Available from: https://doi.org/10.1016/j.physd.2019.132306
- Al-Selwi SM, Hassan MF, Abdulkadir SJ, Muneer A, Sumiea EH, Alqushaibi A, et al. RNN-LSTM: From applications to modeling techniques and beyond—Systematic review. J King Saud Univ Comput Inf Sci. 2024:102068. Available from: https://doi.org/10.1016/j.jksuci.2024.102068
- Shewalkar A, Nyavanandi D, Ludwig SA. Performance evaluation of deep neural networks applied to speech recognition: RNN, LSTM and GRU. J Artif Intell Soft Comput Res. 2019;9:235-245. Available from: http://dx.doi.org/10.2478/jaiscr-2019-0006
- Gao R, Hou X, Qin J, Chen J, Liu L, Zhu F, et al. Zero-VAE-GAN: Generating unseen features for generalized and transductive zero-shot learning. IEEE Trans Image Process. 2020;29:3665-3680. Available from: https://doi.org/10.1109/tip.2020.2964429
- Tian C, Ma Y, Cammon J, Fang F, Zhang Y, Meng M. Dual-encoder VAE-GAN with spatiotemporal features for emotional EEG data augmentation. IEEE Trans Neural Syst Rehabil Eng. 2023;31:2018-2027. Available from: https://doi.org/10.1109/tnsre.2023.3266810
- Ibrahim BI, Nicolae DC, Khan A, Ali SI, Khattak A. VAE-GAN based zero-shot outlier detection. In: Proceedings of the 2020 4th international symposium on computer science and intelligent control. 2020. Available from: https://doi.org/10.1145/3440084.3441180
- Mukesh K, Ippatapu VS, Chereddy S, Anbazhagan E, Oviya IR. A variational autoencoder general adversarial networks (VAE-GAN) based model for ligand designing. In: International Conference on Innovative Computing and Communications: Proceedings of ICICC 2022, Volume 1. Singapore: Springer Nature Singapore; 2022. Available from: https://www.amrita.edu/publication/a-variationalautoencoder-general-adversarial-networks-vae-gan-based-model-for-ligand-designing/
- Elsken T, Metzen JH, Hutter F. Neural architecture search: A survey. J Mach Learn Res. 2019;20(55):1-21. Available from: https://www.jmlr.org/papers/volume20/18-598/18-598.pdf
- Ren P, Xiao Y, Chang X, Huang PY, Li Z, Chen X, et al. A comprehensive survey of neural architecture search: Challenges and solutions. ACM Comput Surv. 2021;54(4):1-34. Available from: https://arxiv.org/abs/2006.02903
- Chitty-Venkata KT, Somani AK. Neural architecture search survey: A hardware perspective. ACM Comput Surv. 2022;55(4):1-36. Available from: http://dx.doi.org/10.1145/3524500
- Li L, Talwalkar A. Random search and reproducibility for neural architecture search. In: Uncertainty in artificial intelligence. PMLR; 2020. p. 367-377. Available from: https://arxiv.org/abs/1902.07638
- Lindauer M, Hutter F. Best practices for scientific research on neural architecture search. J Mach Learn Res. 2020;21(243):1-18. Available from: https://doi.org/10.48550/arXiv.1909.02453
- Mousavi SS, Schukat M, Howley E. Deep reinforcement learning: an overview. In: Proceedings of SAI Intelligent Systems Conference (IntelliSys) 2016: Volume 2. Springer International Publishing; 2018. Available from: https://doi.org/10.48550/arXiv.1806.08894
- Ding Z, Huang Y, Yuan H, Dong H. Introduction to reinforcement learning. Deep reinforcement learning: fundamentals, research and applications. 2020:47-123. Available from: http://dx.doi.org/10.1007/978-981-15-4095-0_2
- Mosavi A, Faghan Y, Ghamisi P, Duan P, Ardabili SF, Salwana E, et al. Comprehensive review of deep reinforcement learning methods and applications in economics. Mathematics. 2020;8(10):1640. Available from: https://doi.org/10.3390/math8101640
- Barto AG. Reinforcement learning: An introduction. SIAM Rev. 2021;6(2):423.
- Gahar RM, Arfaoui O, Hidri MS, Hadj-Alouane NB. A distributed approach for high-dimensionality heterogeneous data reduction. IEEE Access. 2019;7:151006-151022. Available from: https://ieeexplore.ieee.org/stamp/stamp.jsp?arnumber=8861035
- Yilmaz Y, Aktukmak M, Hero AO. Multimodal data fusion in high-dimensional heterogeneous datasets via generative models. IEEE Trans Signal Process. 2021;69:5175-5188. Available from: https://doi.org/10.48550/arXiv.2108.12445
- Pölsterl S, Conjeti S, Navab N, Katouzian A. Survival analysis for high-dimensional, heterogeneous medical data: Exploring feature extraction as an alternative to feature selection. Artif Intell Med. 2016;72:1-11. Available from: https://doi.org/10.1016/j.artmed.2016.07.004
- Rabiee M, Mirhashemi M, Pangburn MS, Piri S, Delen D. Towards explainable artificial intelligence through expert-augmented supervised feature selection. Decis Support Syst. 2024;181:114214. Available from: https://doi.org/10.1016/j.dss.2024.114214
- Aguilar-Ruiz JS. Class-specific feature selection for classification explainability. ArXiv Preprint. 2024. Available from: https://doi.org/10.48550/arXiv.2411.01204
- Woo S, Park J, Lee JY, Kweon IS. CBAM: Convolutional block attention module. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2018. Available from: https://doi.org/10.48550/arXiv.1807.06521
- Vora S, Lang AH, Helou B, Beijbom O. PointPainting: Sequential fusion for 3D object detection. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR). 2020. p. 4604-4612. Available from: https://doi.org/10.48550/arXiv.1911.10150
- Lin Z, Akin H, Rao R, Hie B, Zhu Z, Lu W, et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science. 2023;379:1123-1130. Available from: https://doi.org/10.1126/science.ade2574
- Rostami M, Oussalah M. A novel explainable COVID-19 diagnosis method by integration of feature selection with random forest. Inform Med Unlocked. 2022;30:100941. Available from: https://doi.org/10.1016/j.imu.2022.100941
- Panhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng. 2016;63(7):1455-1462. Available from: https://doi.org/10.1109/tbme.2015.2496264
- Li X. Federated feature learning for mammography diagnosis with privacy preservation. Nat Digit Med. 2022;5:123.
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Proc IEEE Int Conf Comput Vis (ICCV). 2017;618-626. Available from: https://doi.org/10.48550/arXiv.1610.02391
- Wang H. Interpretable AI for lung nodule malignancy prediction in low-dose CT. Nat Med. 2023;29(6):1430-1438.
- Chen T, Kornblith S, Norouzi M, Hinton G. A simple framework for contrastive learning of visual representations. Proc Int Conf Mach Learn (ICML). 2020;119:1597-1607. Available from: https://proceedings.mlr.press/v119/chen20j.html
- Howard A, Sandler M, Chu G, Chen LC, Chen B, Tan M, et al. Searching for MobileNetV3. Proc IEEE/CVF Int Conf Comput Vis (ICCV). 2019;1314-1324. Available from: https://openaccess.thecvf.com/content_ICCV_2019/html/Howard_Searching_for_MobileNetV3_ICCV_2019_paper.html
- Zhang Y. Wavelet-CNN for mechanical fault diagnosis under noisy environments. Mech Syst Signal Process. 2021;152:107413.
- Gupta A. Multimodal sensor fusion for predictive maintenance in Industry 4.0. IEEE Trans Ind Inform. 2023;19(7):4321-4332.
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583-589. Available from: https://www.nature.com/articles/s41586-021-03819-2
Figures:
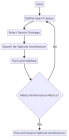
Figure 1
Similar Articles
-
Survey of Advanced Image Fusion Techniques for Enhanced Visualization in Cardiovascular Diagnosis and TreatmentGargi J Trivedi*. Survey of Advanced Image Fusion Techniques for Enhanced Visualization in Cardiovascular Diagnosis and Treatment. . 2025 doi: 10.29328/journal.jcmei.1001034; 9: 001-009
-
Feature Processing Methods: Recent Advances and Future TrendsShiying Bai,Lufeng Bai*. Feature Processing Methods: Recent Advances and Future Trends. . 2025 doi: 10.29328/journal.jcmei.1001035; 9: 010-014
Recently Viewed
-
Cystoid Macular Oedema Secondary to Bimatoprost in a Patient with Primary Open Angle GlaucomaKonstantinos Kyratzoglou*,Katie Morton. Cystoid Macular Oedema Secondary to Bimatoprost in a Patient with Primary Open Angle Glaucoma. Int J Clin Exp Ophthalmol. 2025: doi: 10.29328/journal.ijceo.1001059; 9: 001-003
-
Sex after Neurosurgery–Limitations, Recommendations, and the Impact on Patient’s Well-beingMor Levi Rivka*, Csaba L Dégi. Sex after Neurosurgery–Limitations, Recommendations, and the Impact on Patient’s Well-being. J Neurosci Neurol Disord. 2024: doi: 10.29328/journal.jnnd.1001099; 8: 064-068
-
Physiotherapy Undergraduate Students’ Perception About Clinical Education; A Qualitative StudyPravakar Timalsina*,Bimika Khadgi. Physiotherapy Undergraduate Students’ Perception About Clinical Education; A Qualitative Study. J Nov Physiother Rehabil. 2024: doi: 10.29328/journal.jnpr.1001063; 8: 043-052
-
Clinical Significance of Anterograde Angiography for Preoperative Evaluation in Patients with Varicose VeinsYi Liu,Dong Liu#,Junchen Li#,Tianqing Yao,Yincheng Ran,Ke Tian,Haonan Zhou,Lei Zhou,Zhumin Cao*,Kai Deng*. Clinical Significance of Anterograde Angiography for Preoperative Evaluation in Patients with Varicose Veins. J Radiol Oncol. 2025: doi: 10.29328/journal.jro.1001073; 9: 001-006
-
Regional Anesthesia Challenges in a Pregnant Patient with VACTERL Association: A Case ReportUzma Khanam*,Abid,Bhagyashri V Kumbar. Regional Anesthesia Challenges in a Pregnant Patient with VACTERL Association: A Case Report. Int J Clin Anesth Res. 2025: doi: 10.29328/journal.ijcar.1001027; 9: 010-012
Most Viewed
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."